Protein:DACH1 |
Protein Summary |
 Gene summary Gene summary |
| Gene name: DACH1 | ASpdb.0 ID: 1602 | Gene | Gene symbol | DACH1 | Gene ID | 1602 |
| Gene name | dachshund family transcription factor 1 |
| Synonyms | DACH |
| Cytomap | 13q21.33 |
| Type of gene | protein-coding |
| Description | dachshund homolog 1dac homolog |
| Modification date | 20240408 |
| UniProtAcc | Q9UI36 |
 Gene ontology of this gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of this gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Gene | DACH1 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 20956529 |
| Gene | DACH1 | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | 20956529 |
| Gene | DACH1 | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | 20956529 |
| Gene | DACH1 | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | 20956529 |
| Gene | DACH1 | GO:0005654 | nucleoplasm | - |
| Gene | DACH1 | GO:0005794 | Golgi apparatus | - |
| Gene | DACH1 | GO:0005829 | cytosol | - |
| Gene | DACH1 | GO:0016607 | nuclear speck | - |
| Gene | DACH1 | GO:0030336 | negative regulation of cell migration | 20956529 |
| Gene | DACH1 | GO:2000279 | negative regulation of DNA biosynthetic process | 16980615 |
AS Summary |
 Information of the canonical protein with experimentally identified structure from PDB (2023). Information of the canonical protein with experimentally identified structure from PDB (2023). |
| UniProt Acc | File name | PDB ID | Method | Resolution | Chain | Start | End |
| Q9UI36-1 | Q9UI36-1_1l8r_A.pdb | 1L8R | X-ray | 1.65 | A | 184 | 282 |
 ASpdb's canonical and alternatively spliced isoform information. ASpdb's canonical and alternatively spliced isoform information. |
| accession_id | gene_name | canonical_id | alternative_id | canonical_length | alternative_length | canonical_start | canonical_end | type | originalSEQ | variationSEQ | alternative_start | alternative_end |
| Q9UI36 | DACH1 | Q9UI36-1 | Q9UI36-2 | 758 | 706 | 376 | 427 | Deletion | none | none | 375 | 375 |
| Q9UI36 | DACH1 | Q9UI36-1 | Q9UI36-3 | 758 | 558 | 376 | 427 | Deletion | none | none | 375 | 375 |
| Q9UI36 | DACH1 | Q9UI36-1 | Q9UI36-3 | 758 | 558 | 428 | 575 | Deletion | none | none | 375 | 375 |
| Q9UI36 | DACH1 | Q9UI36-1 | Q9UI36-4 | 758 | 504 | 322 | 575 | Deletion | none | none | 321 | 321 |
 Multiple sequence alignment of our canonical and alternatively spliced DACH1 Multiple sequence alignment of our canonical and alternatively spliced DACH1 |
 Matched gene isoform IDs with Ensembl and RefSeq of our canonical and alternative spliced genes of DACH1 Matched gene isoform IDs with Ensembl and RefSeq of our canonical and alternative spliced genes of DACH1 |
| UniProt-id | ENSG | ENST | ENSP |
| Q9UI36-1 | ENSG00000276644.5 | ENST00000619232.2 | ENSP00000482797.1 |
| Q9UI36-2 | ENSG00000276644.5 | ENST00000613252.5 | ENSP00000482245.1 |
| UniProt-id | NM ID | NP ID |
| Q9UI36-2 | NM_080759.5 | NP_542937.3 |
| Q9UI36-3 | NM_080760.5 | NP_542938.3 |
| Q9UI36-4 | NM_004392.6 | NP_004383.4 |
 Amino acid sequences of our canonical and alternatively spliced DACH1 Amino acid sequences of our canonical and alternatively spliced DACH1 |
| accession_id | Protein sequence |
| Q9UI36-1 | MAVPAALIPPTQLVPPQPPISTSASSSGTTTSTSSATSSPAPSIGPPASSGPTLFRPEPIASAAAAAATVTSTGGGGGGGGSGGGGGSSG NGGGGGGGGGGSNCNPNLAAASNGSGGGGGGISAGGGVASSTPINASTGSSSSSSSSSSSSSSSSSSSSSSSSCGPLPGKPVYSTPSPVE NTPQNNECKMVDLRGAKVASFTVEGCELICLPQAFDLFLKHLVGGLHTVYTKLKRLEITPVVCNVEQVRILRGLGAIQPGVNRCKLISRK DFETLYNDCTNASSRPGRPPKRTQSVTSPENSHIMPHSVPGLMSPGIIPPTGLTAAAAAAAAATNAAIAEAMKVKKIKLEAMSNYHASNN QHGADSENGDMNSSVGSSDGSWDKETLPSSPSQGPQASITHPRMPGARSLPLSHPLNHLQQSHLLPNGLELPFMMMPHPLIPVSLPPASV TMAMSQMNHLSTIANMAAAAQVQSPPSRVETSVIKERVPDSPSPAPSLEEGRRPGSHPSSHRSSSVSSSPARTESSSDRIPVHQNGLSMN QMLMGLSPNVLPGPKEGDLAGHDMGHESKRMHIEKDETPLSTPTARDSLDKLSLTGHGQPLPPGFPSPFLFPDGLSSIETLLTNIQGLLK VAIDNARAQEKQVQLEKTELKMDFLRERELRETLEKQLAMEQKNRAIVQKRLKKEKKAKRKLQEALEFETKRREQAEQTLKQAASTDSLR |
| Q9UI36-2 | MAVPAALIPPTQLVPPQPPISTSASSSGTTTSTSSATSSPAPSIGPPASSGPTLFRPEPIASAAAAAATVTSTGGGGGGGGSGGGGGSSG NGGGGGGGGGGSNCNPNLAAASNGSGGGGGGISAGGGVASSTPINASTGSSSSSSSSSSSSSSSSSSSSSSSSCGPLPGKPVYSTPSPVE NTPQNNECKMVDLRGAKVASFTVEGCELICLPQAFDLFLKHLVGGLHTVYTKLKRLEITPVVCNVEQVRILRGLGAIQPGVNRCKLISRK DFETLYNDCTNASSRPGRPPKRTQSVTSPENSHIMPHSVPGLMSPGIIPPTGLTAAAAAAAAATNAAIAEAMKVKKIKLEAMSNYHASNN QHGADSENGDMNSSVGLELPFMMMPHPLIPVSLPPASVTMAMSQMNHLSTIANMAAAAQVQSPPSRVETSVIKERVPDSPSPAPSLEEGR RPGSHPSSHRSSSVSSSPARTESSSDRIPVHQNGLSMNQMLMGLSPNVLPGPKEGDLAGHDMGHESKRMHIEKDETPLSTPTARDSLDKL SLTGHGQPLPPGFPSPFLFPDGLSSIETLLTNIQGLLKVAIDNARAQEKQVQLEKTELKMDFLRERELRETLEKQLAMEQKNRAIVQKRL |
| Q9UI36-3 | MAVPAALIPPTQLVPPQPPISTSASSSGTTTSTSSATSSPAPSIGPPASSGPTLFRPEPIASAAAAAATVTSTGGGGGGGGSGGGGGSSG NGGGGGGGGGGSNCNPNLAAASNGSGGGGGGISAGGGVASSTPINASTGSSSSSSSSSSSSSSSSSSSSSSSSCGPLPGKPVYSTPSPVE NTPQNNECKMVDLRGAKVASFTVEGCELICLPQAFDLFLKHLVGGLHTVYTKLKRLEITPVVCNVEQVRILRGLGAIQPGVNRCKLISRK DFETLYNDCTNASSRPGRPPKRTQSVTSPENSHIMPHSVPGLMSPGIIPPTGLTAAAAAAAAATNAAIAEAMKVKKIKLEAMSNYHASNN QHGADSENGDMNSSVDETPLSTPTARDSLDKLSLTGHGQPLPPGFPSPFLFPDGLSSIETLLTNIQGLLKVAIDNARAQEKQVQLEKTEL KMDFLRERELRETLEKQLAMEQKNRAIVQKRLKKEKKAKRKLQEALEFETKRREQAEQTLKQAASTDSLRVLNDSLTPEIEADRSGGRTD |
| Q9UI36-4 | MAVPAALIPPTQLVPPQPPISTSASSSGTTTSTSSATSSPAPSIGPPASSGPTLFRPEPIASAAAAAATVTSTGGGGGGGGSGGGGGSSG NGGGGGGGGGGSNCNPNLAAASNGSGGGGGGISAGGGVASSTPINASTGSSSSSSSSSSSSSSSSSSSSSSSSCGPLPGKPVYSTPSPVE NTPQNNECKMVDLRGAKVASFTVEGCELICLPQAFDLFLKHLVGGLHTVYTKLKRLEITPVVCNVEQVRILRGLGAIQPGVNRCKLISRK DFETLYNDCTNASSRPGRPPKRTQSVTSPENSHIMPHSVPGLMSPGIIPPTDETPLSTPTARDSLDKLSLTGHGQPLPPGFPSPFLFPDG LSSIETLLTNIQGLLKVAIDNARAQEKQVQLEKTELKMDFLRERELRETLEKQLAMEQKNRAIVQKRLKKEKKAKRKLQEALEFETKRRE |
Protein Functional Features |
 Main function of this protein. (from UniProt) Main function of this protein. (from UniProt) |
| DACH1 (go to UniProt):Q9UI36 |
 Retention analysis result of protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Accession_id | Subsection | Start | End | Funcitonal feature | Splicing information |
| Q9UI36 | Region | 189 | 384 | Note=Interaction with SIX6 and HDAC3;Ontology_term=ECO:0000250;evidence=ECO:0000250 | Type=Deletion;Start=376;End=427 |
| Q9UI36 | Region | 189 | 384 | Note=Interaction with SIX6 and HDAC3;Ontology_term=ECO:0000250;evidence=ECO:0000250 | Type=Deletion;Start=376;End=427 |
| Q9UI36 | Region | 189 | 384 | Note=Interaction with SIX6 and HDAC3;Ontology_term=ECO:0000250;evidence=ECO:0000250 | Type=Deletion;Start=322;End=575 |
| Q9UI36 | Region | 358 | 414 | Note=Disordered;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=376;End=427 |
| Q9UI36 | Region | 358 | 414 | Note=Disordered;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=376;End=427 |
| Q9UI36 | Region | 358 | 414 | Note=Disordered;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=322;End=575 |
| Q9UI36 | Region | 474 | 532 | Note=Disordered;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=428;End=575 |
| Q9UI36 | Region | 474 | 532 | Note=Disordered;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=322;End=575 |
| Q9UI36 | Region | 544 | 564 | Note=Disordered;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=428;End=575 |
| Q9UI36 | Region | 544 | 564 | Note=Disordered;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=322;End=575 |
| Q9UI36 | Compositional bias | 358 | 399 | Note=Polar residues;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=376;End=427 |
| Q9UI36 | Compositional bias | 358 | 399 | Note=Polar residues;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=376;End=427 |
| Q9UI36 | Compositional bias | 358 | 399 | Note=Polar residues;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=322;End=575 |
| Q9UI36 | Compositional bias | 508 | 532 | Note=Polar residues;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=428;End=575 |
| Q9UI36 | Compositional bias | 508 | 532 | Note=Polar residues;Ontology_term=ECO:0000256;evidence=ECO:0000256|SAM:MobiDB-lite | Type=Deletion;Start=322;End=575 |
Gene Isoform Structures and Expression Levels for DACH1 |
 Gene structures of our canonical and alternative spliced genes of DACH1 Gene structures of our canonical and alternative spliced genes of DACH1* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Expression levels of gene isoforms across GTEx. Expression levels of gene isoforms across GTEx. |
 Expression levels of gene isoforms across TCGA. Expression levels of gene isoforms across TCGA. |
Protein Structures |
 PDB and CIF files of the predicted protein structures PDB and CIF files of the predicted protein structures * Here we show the 3D structure of the proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| 3D view using mol* of Q9UI36-1 |
| 3D view using mol* of Q9UI36-2 |
| 3D view using mol* of Q9UI36-3 |
| 3D view using mol* of Q9UI36-4 |
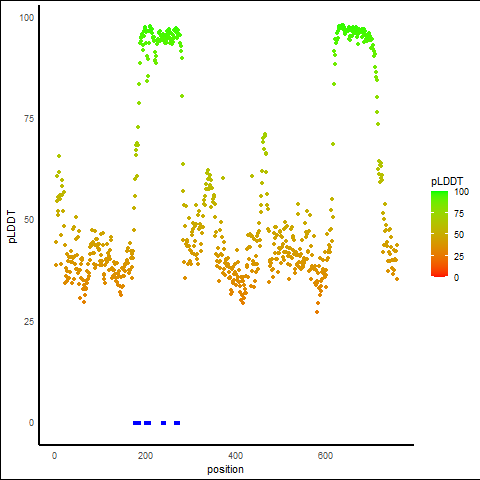
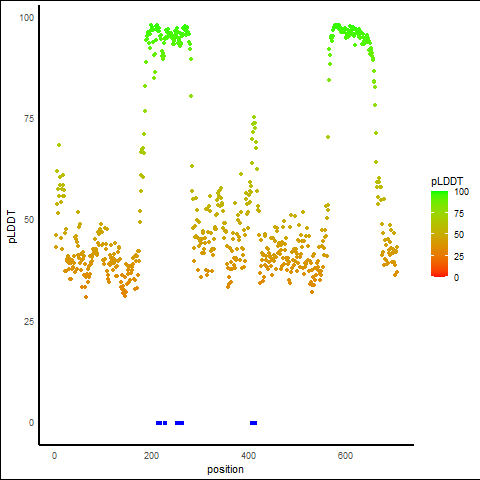
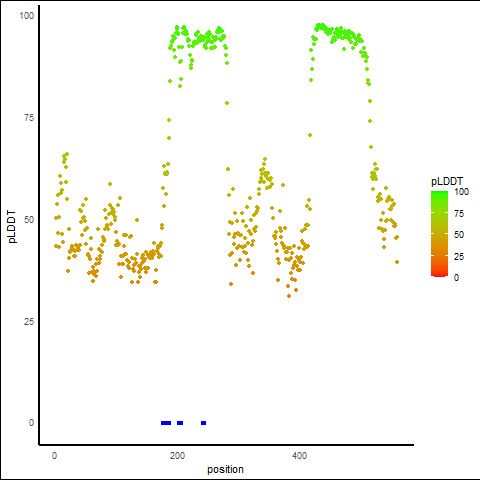
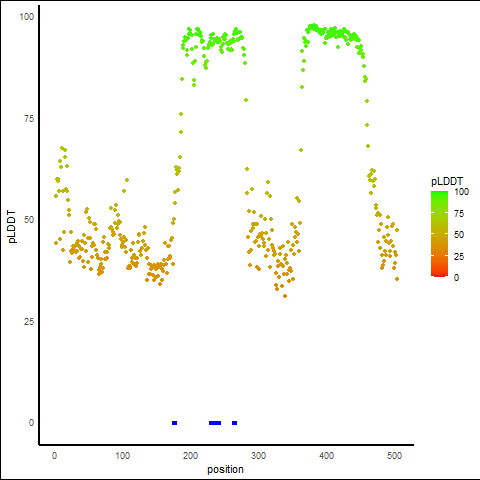
pLDDT Score Distribution |
 pLDDT score distribution of the predicted protein structures from AlphaFold2 pLDDT score distribution of the predicted protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
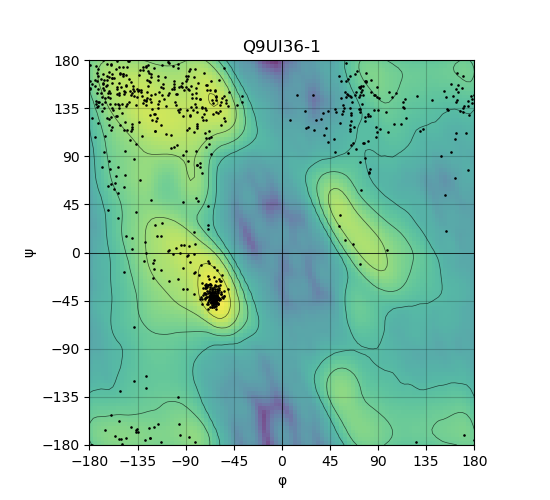
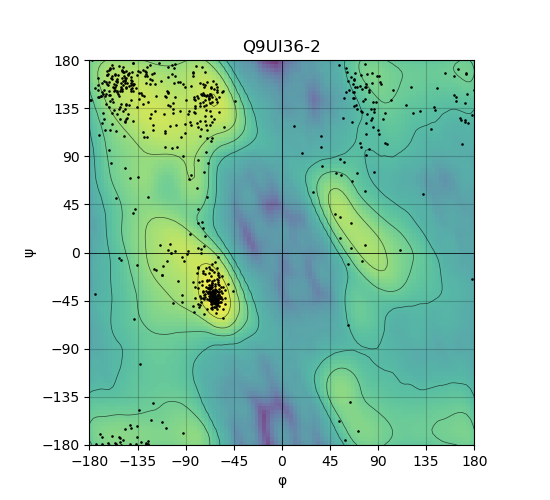
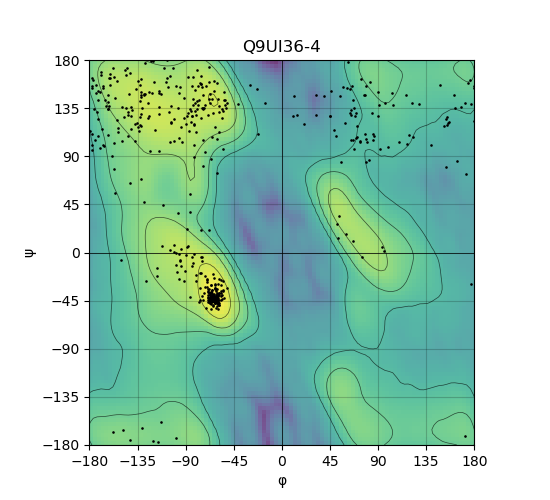
Ramachandran Plot of Protein Structures |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this protein peptide. |
| Ramachandran plot of Q9UI36-1 |
 |
| Ramachandran plot of Q9UI36-2 |
 |
| Ramachandran plot of Q9UI36-4 |
 |
Potential Active Site Information |
 The potential binding sites of these proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these proteins were identified using SiteMap, a module of the Schrodinger suite. |
| UniProt-id | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| Q9UI36-1 | 0.719 | 54 | 0.673 | 156.065 | 0.727 | 0.568 | 0.778 | 0.112 | 1.101 | 0.102 | 0.642 | 176,177,178,179,180,182,184,185,202,203,204,205,20 8,238,239,240,241,242,267,268,271 |
| Q9UI36-2 | 0.776 | 56 | 0.727 | 169.785 | 0.665 | 0.639 | 0.826 | 0.21 | 1.102 | 0.19 | 0.996 | 212,213,216,217,226,252,253,255,256,257,258,259,26 1,406,407,409,410,413 |
| Q9UI36-3 | 0.636 | 43 | 0.61 | 102.9 | 0.777 | 0.506 | 0.651 | 0.137 | 0.951 | 0.144 | 1.057 | 176,177,178,179,180,182,184,185,202,203,204,205,24 1,242 |
| Q9UI36-4 | 0.612 | 26 | 0.544 | 104.958 | 0.72 | 0.635 | 0.888 | 0.289 | 0.971 | 0.297 | 0.813 | 175,176,230,233,234,238,239,240,241,242,263,265
|
Protein Structure and Feature Comparision |
 Protein Structure Comparision Using Template Modeling Scores (TM-score). Protein Structure Comparision Using Template Modeling Scores (TM-score). |
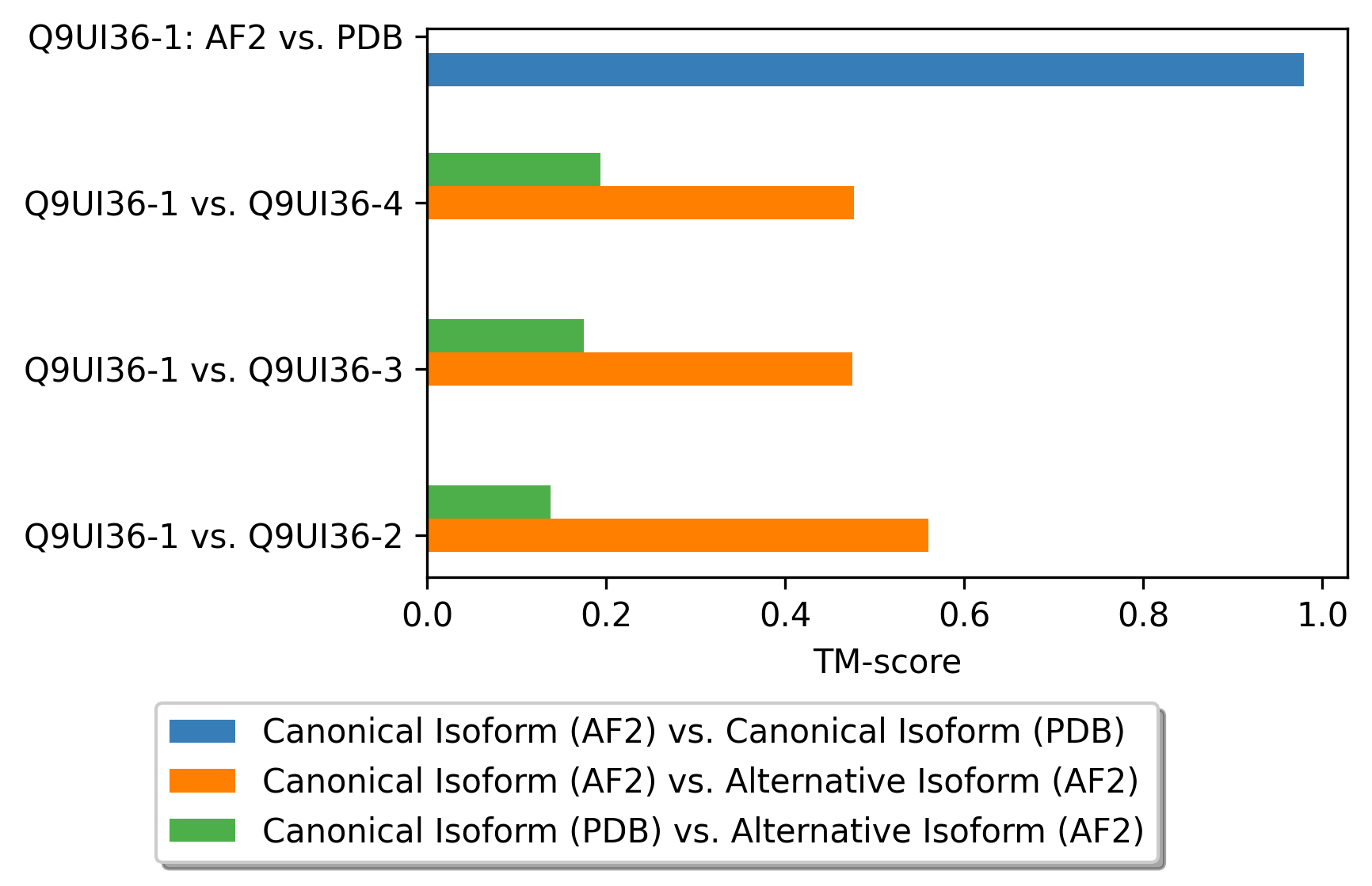 |
 Protein Structure Comparision Visualization with mol*. between Canonical predicted structure (AF2)(orange) vs Canonical validated structure (PDB)(green) Protein Structure Comparision Visualization with mol*. between Canonical predicted structure (AF2)(orange) vs Canonical validated structure (PDB)(green) |
| 3D view using mol* of Q9UI36-1_Q9UI36-1_1l8r_A.pdb |
 Protein Structure Comparision Visualization with mol*. between Canonical validated structure (PDB)(orange) vs Alternative predicted structure (AF2)(green) Protein Structure Comparision Visualization with mol*. between Canonical validated structure (PDB)(orange) vs Alternative predicted structure (AF2)(green) |
| 3D view using mol* of Q9UI36-1_1l8r_A_Q9UI36-2.pdb |
| 3D view using mol* of Q9UI36-1_1l8r_A_Q9UI36-3.pdb |
| 3D view using mol* of Q9UI36-1_1l8r_A_Q9UI36-4.pdb |
 Protein Structure Comparision Visualization with mol*. between Canonical predicted structure (AF2)(orange) vs Alternative predicted structure (AF2)(green) Protein Structure Comparision Visualization with mol*. between Canonical predicted structure (AF2)(orange) vs Alternative predicted structure (AF2)(green) |
| 3D view using mol* of Q9UI36-1_Q9UI36-2.pdb |
| 3D view using mol* of Q9UI36-1_Q9UI36-3.pdb |
| 3D view using mol* of Q9UI36-1_Q9UI36-4.pdb |
 Protein Feature Comparison of the protein sequendary structures among the protiens. Protein Feature Comparison of the protein sequendary structures among the protiens. |
| ./stats/secondary_structure/figure/Q9UI36-1_vs_Q9UI36-2.png |
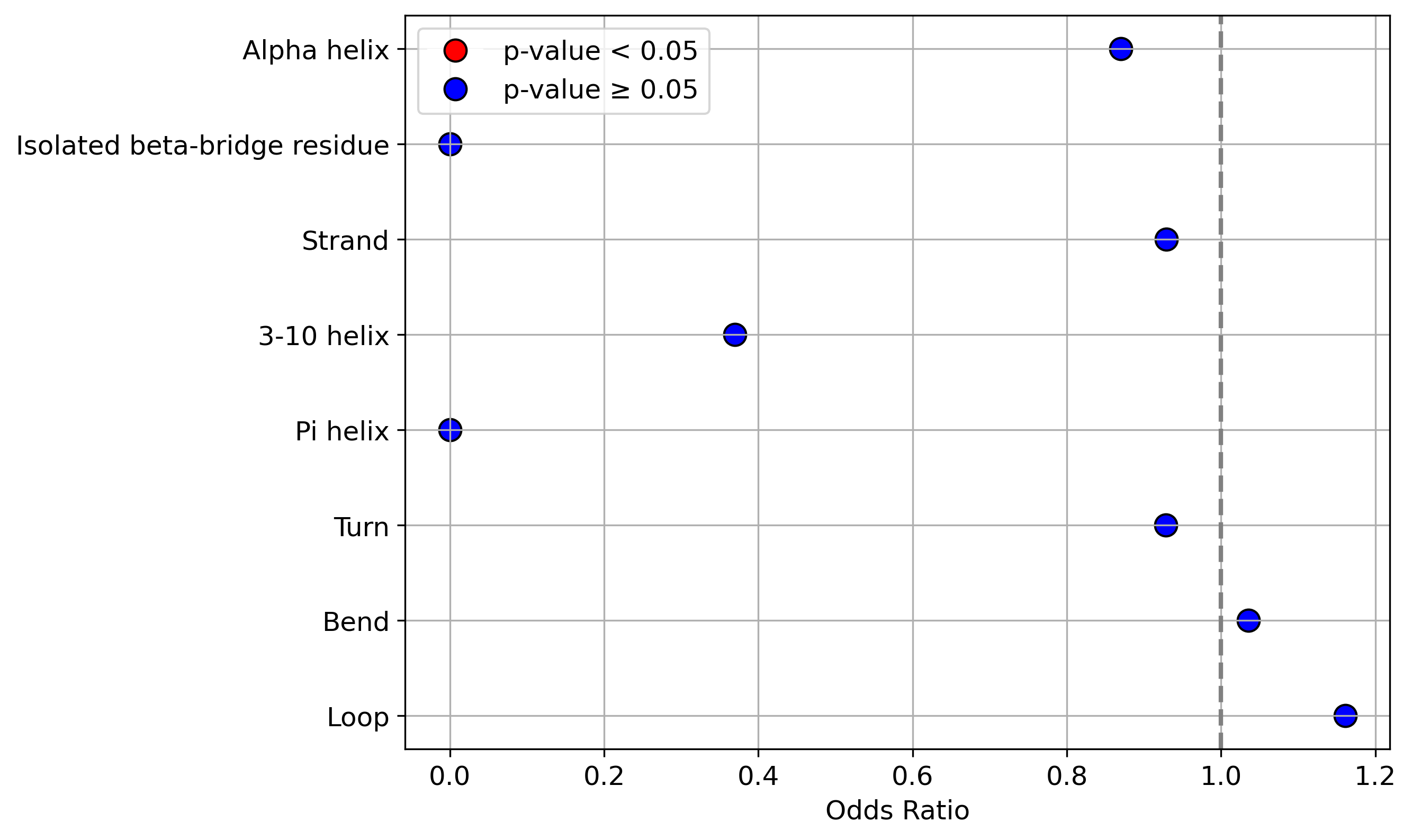 < < |
| ./stats/secondary_structure/figure/Q9UI36-1_vs_Q9UI36-3.png |
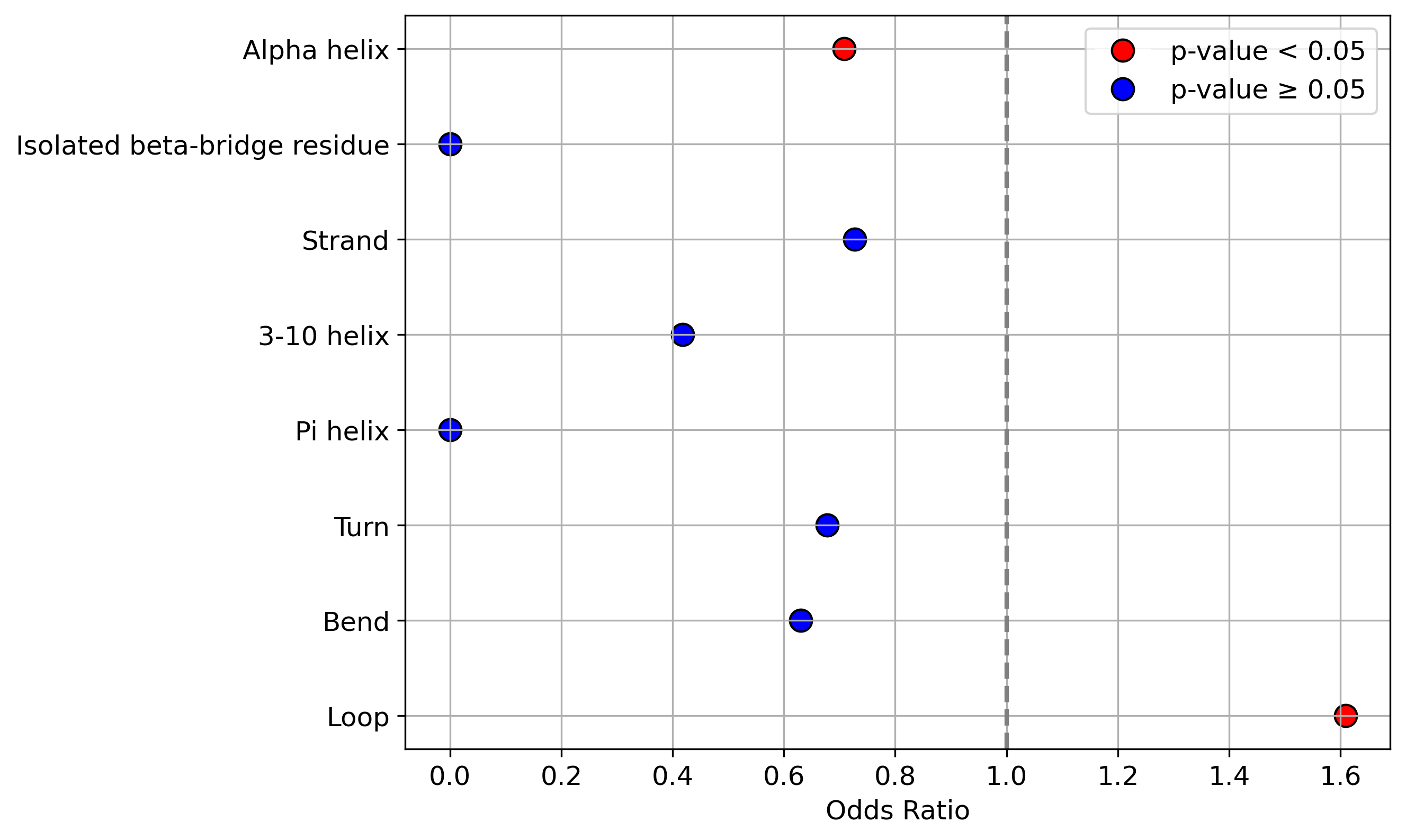 < < |
| ./stats/secondary_structure/figure/Q9UI36-1_vs_Q9UI36-4.png |
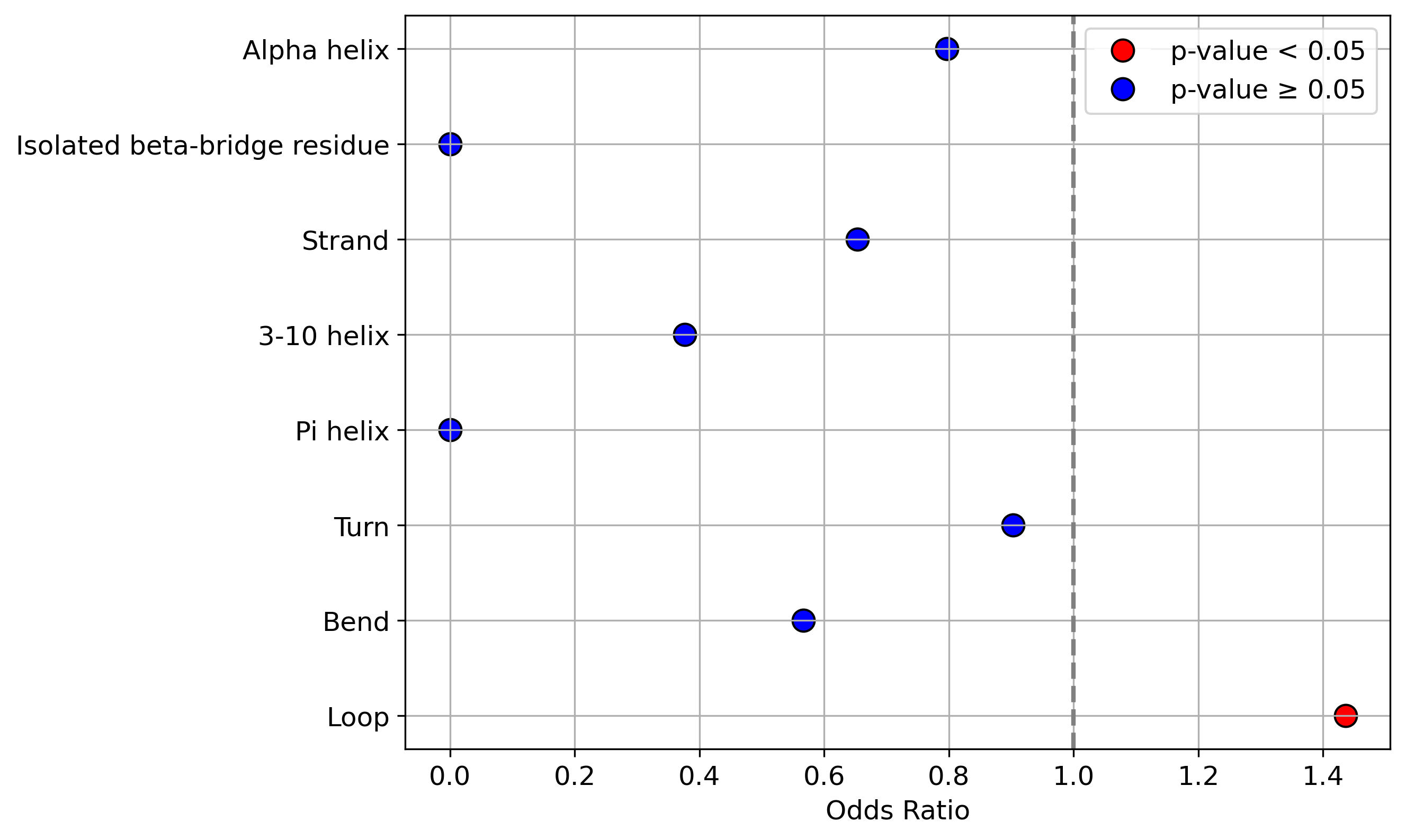 < < |
 Protein Feature Comparison of the relative accessible surface area (ASA) among the protiens. Protein Feature Comparison of the relative accessible surface area (ASA) among the protiens. |
| ./stats/relative_asa/Q9UI36-1_vs_Q9UI36-2.png |
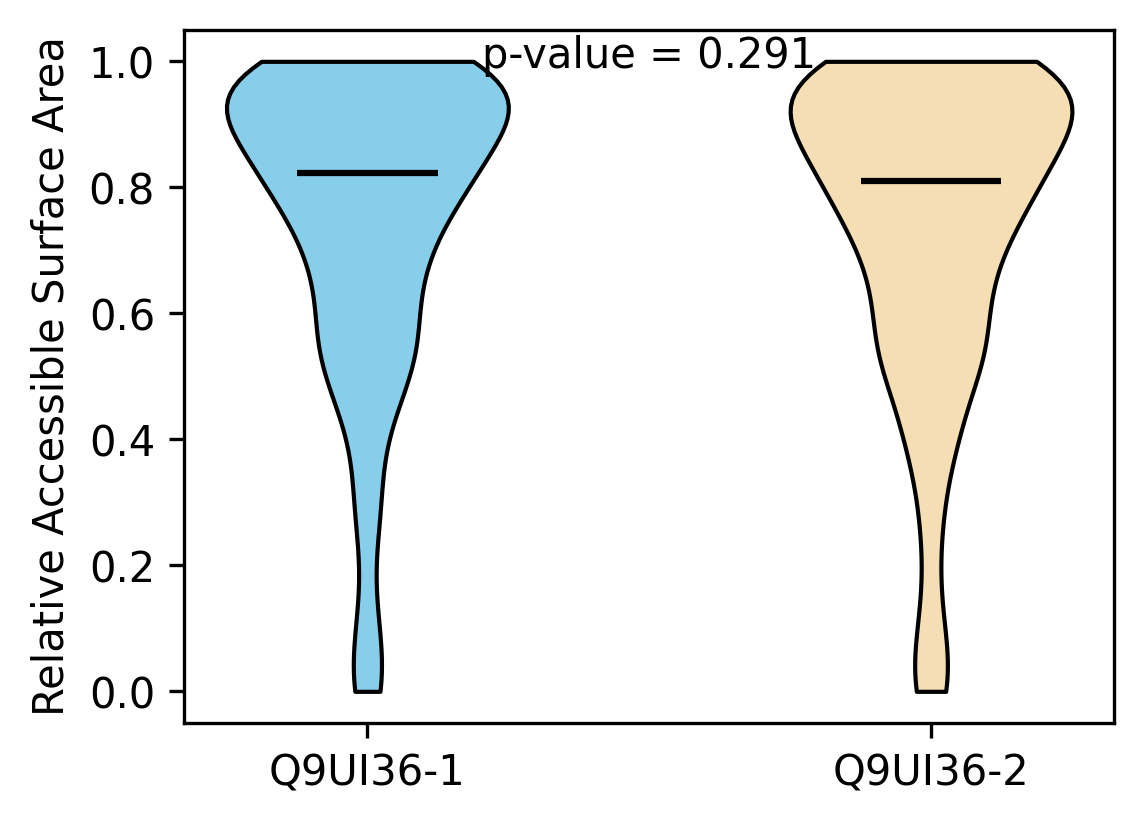 < < |
| ./stats/relative_asa/Q9UI36-1_vs_Q9UI36-3.png |
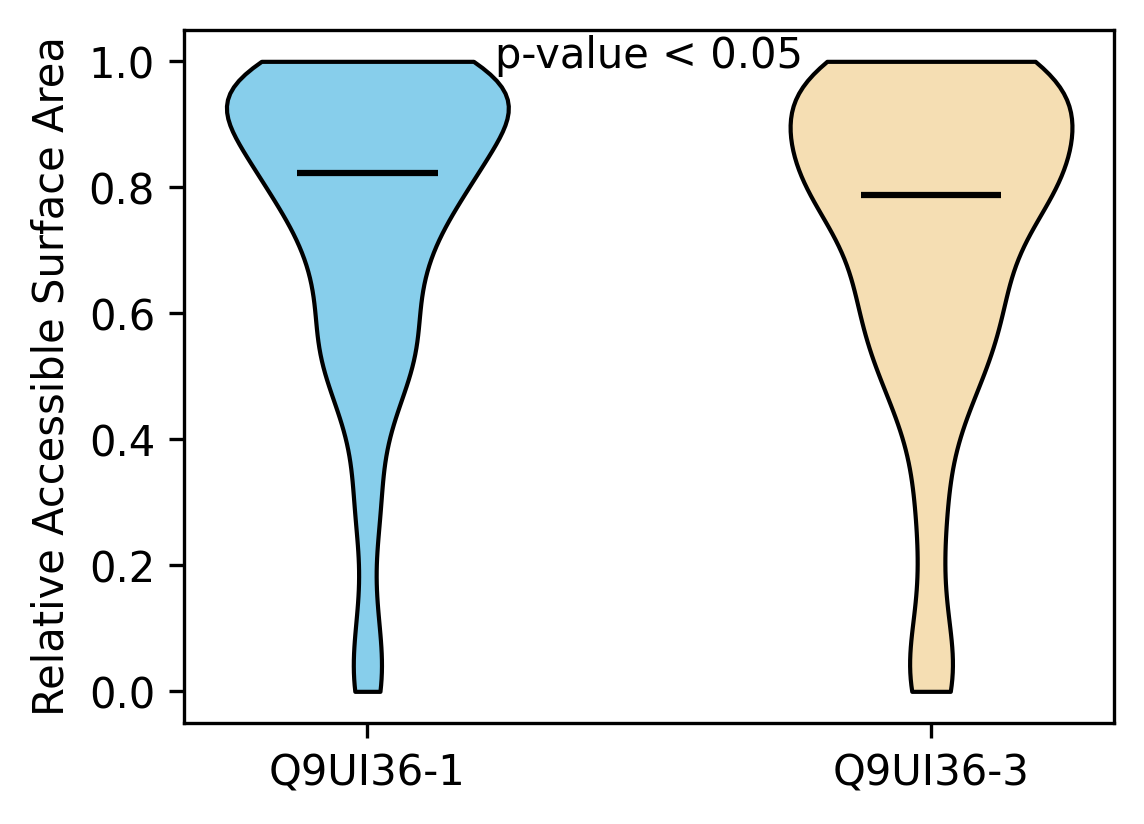 < < |
| ./stats/relative_asa/Q9UI36-1_vs_Q9UI36-4.png |
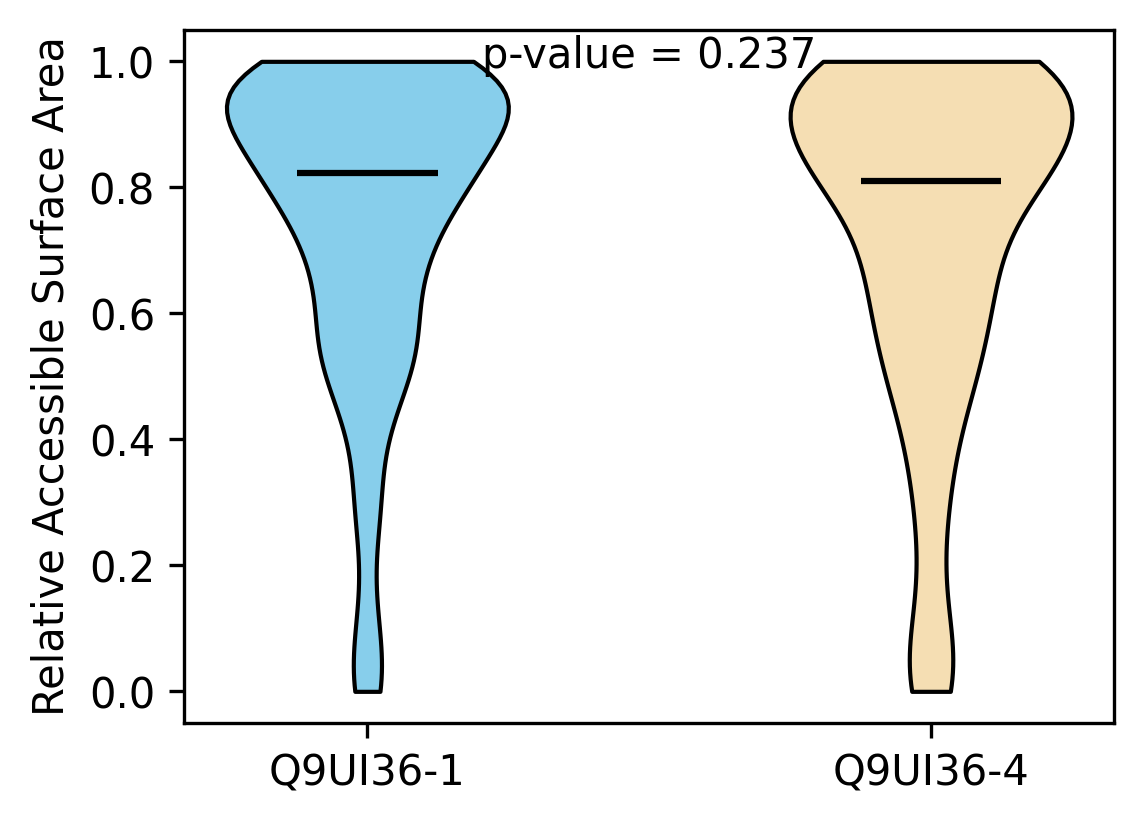 < < |
Protein-Protein Interaction |
 Interactors from UniProt. Interactors from UniProt. |
| Accession_id | Subsection | Start | End | Funcitonal feature | Splicing information |
| Q9UI36 | Region | 189 | 384 | Note=Interaction with SIX6 and HDAC3;Ontology_term=ECO:0000250;evidence=ECO:0000250 | Type=Deletion;Start=376;End=427 |
| Q9UI36 | Region | 189 | 384 | Note=Interaction with SIX6 and HDAC3;Ontology_term=ECO:0000250;evidence=ECO:0000250 | Type=Deletion;Start=376;End=427 |
| Q9UI36 | Region | 189 | 384 | Note=Interaction with SIX6 and HDAC3;Ontology_term=ECO:0000250;evidence=ECO:0000250 | Type=Deletion;Start=322;End=575 |
 Interactors from STRING. Interactors from STRING. |
| Gene name | Interactors |
Related Drugs to DACH1 |
 Drugs targeting this gene/protein. Drugs targeting this gene/protein. (DrugBank) |
| UniProt accession | Gene name | DrugBank ID | Drug name | Drug group | Actions |
Related Diseases to DACH1 |
 Previous studies relating to the alternative splicing of DACH1 and disease information from the MeSH term (PubMed) Previous studies relating to the alternative splicing of DACH1 and disease information from the MeSH term (PubMed) |
| Gene | PMID | Title | Abstract | MeSH ID | MeSH term |
Clinically important variants in DACH1 |
 (ClinVar, 04/20/2024) (ClinVar, 04/20/2024) |
| accession_id | uniprot_id | gene_name | Type | Variant | Clinical_significance |
|
|